SEM via lavaan syntax in jamovi
keywords semlj,jamovi, lavaan, SEM
SEMLj lavaan syntax module of the SEMLj suite for jamovi
0.8.*
Draft version, mistakes may be around
Introduction
The SEMLj module is a jamovi
interface to lavaan R package (Rosseel 2012). The module offers a syntax
interface in which lavaan syntax for the model definition can be passed
to the module. Almost everything you can pass in lavaan, you can pass it
to SEMLj. All the available options are
in the foldable panels below the syntax field. Thus, it is recommended
that the user takes a look at lavaan website for details.
User Interface
When installed, the module sub-modules can be found in the
SEM menu.
Here we need SEM (syntax).
Syntax field
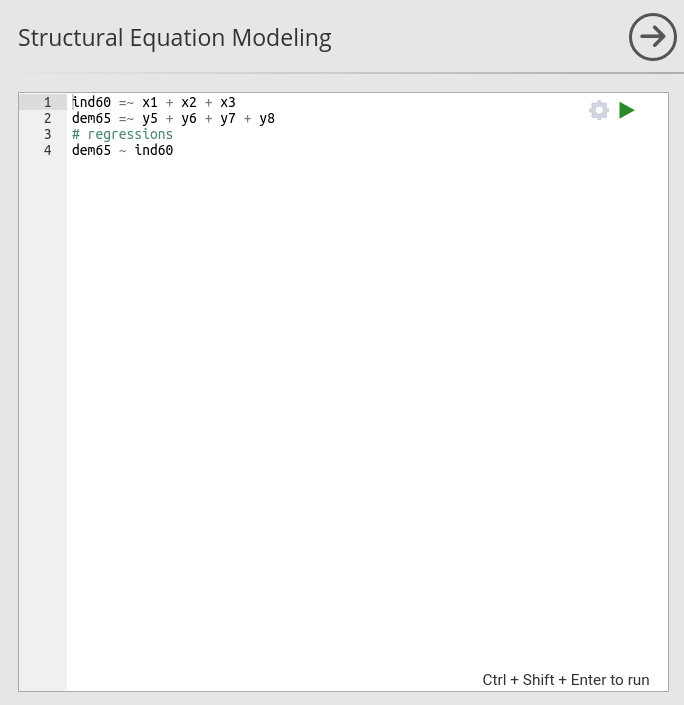
The syntax panel accepts lavaan syntax as described in
lavaan
website.
Note:
The syntax field accepts the lavaan syntax to define a model, not lavaan options or commands. For options, please use the options panels beneath the syntax field.
Warning:
Whereas any syntax you submit will be passed to
lavaan::lavaan(), SEMLj may
be not ready to understand the results and frame them in the output
tables. At the moment, we have tested:
- Path analysis models
- Structural Equation models
- Confirmatory Factor Analysis
- Multigroup Analysis
- Multilevel Models
- Defined parameters (i.e a:=p1^2)
- free and fixing parameters via model syntax (i.e x~~x or a~0*1)
Type of variables
To obtain a standard SEM model, please be sure that the observed
variables are marked as continuous variable in the dataset.
You see a little ruler icon on the left of the variable name,
like this:
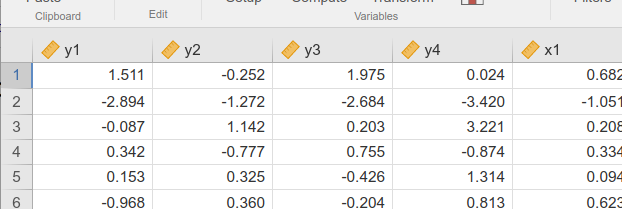
When variables are defined as continuous, they are
considered continuous variables in SEMLj.
Categorical variables can be analyzed if they are defined as
dichotomous or ordinal variable. SEMLj
treats variables as ordered variables if they are defined as
Nominal or Ordinal in the dataset, like
this:
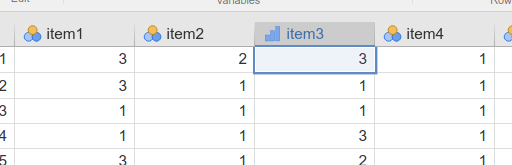
When indicators are ordinal or nominal, Diagonal Weighted Least Squares is used for estimation, with robust standard error. Please refer to lavaan documentation for more details.
Parameters labels
Any coefficient can be labeled in the syntax following
lavaan syntax standard:
This would attach the labels a1, a2 and
a3 to the corresponding coefficient.
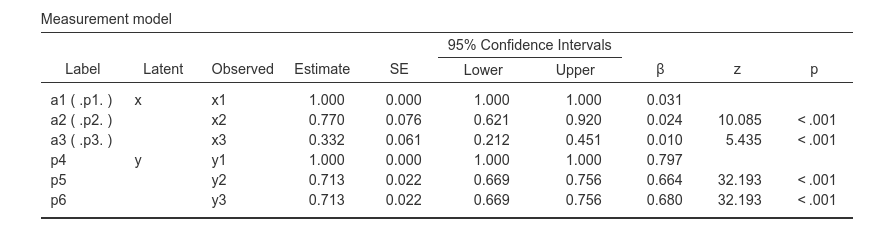
SEMLj automatically assigns a label
to any coefficient, naming them pN, where N is
an increasing integer. So, for y1, y2 and
y3 in the example, a label of p4,
p5 and p6 are assigned. User defined and
automatically assigned labels can be used in the syntax in the same way.
For instance:
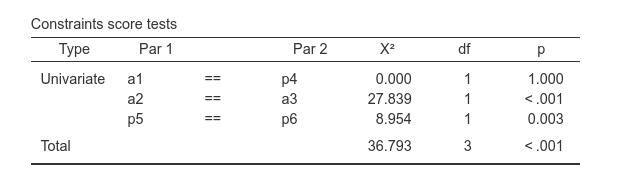
produced the desired constraints and relative tests.
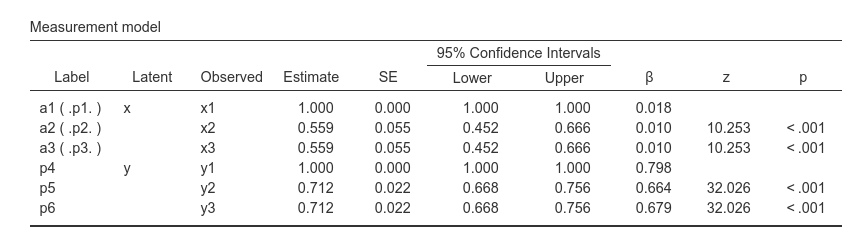
For used defined labels, the tables show also the internal automatic
label uniquely assigned to the coefficients, with dots.
a1 (.p1.) means the coefficient labelled by the user as
a1 but internally referred to as .p1.. This
additional information regarding the internal label is usefull in some
cases of equality constraints.
Consider this syntax:
This asks for a latent variable scaled at y1, but with
the loading of y2,y3 and y4
estimated as equal.
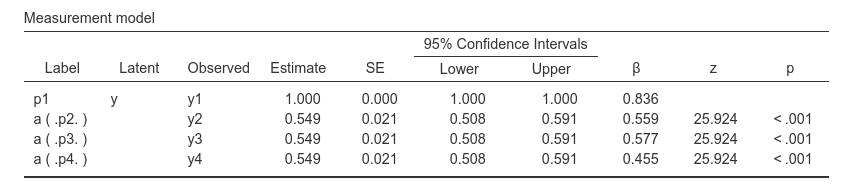
This also produces the tests for the pairwise comparisons of the coefficients.
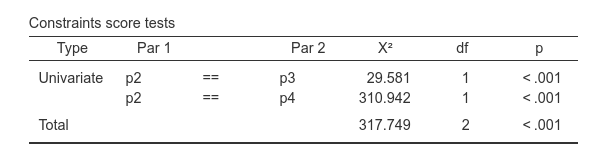
By comparing the uniquely assigned label (p2,
p3, p4), we know which comparison is
tested.
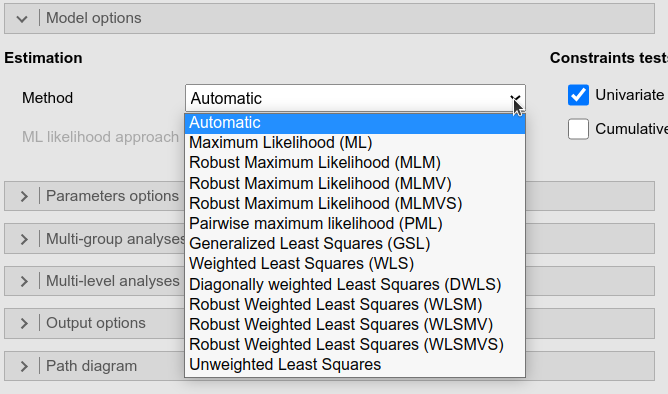
The Estimation Method allows choosing the
right estimation method available for a model. The Automatic option (default) lets
lavaan choose the right method depending to the type of
model one is estimating.
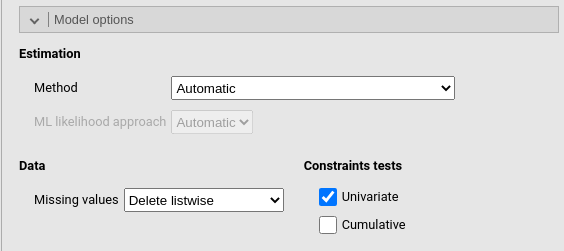
Constraints tests computes the \(\chi^2\) test for contrainted coefficients (if any). In case one has more than one contraints, each constraint is tested uniquely (if Univariate is flagged) and in addition to the previous ones (if Cumulative is flagged).
Under Data one can decide how to handle missing values.
The options for this panel are are:
| Method |
The estimator to be used (cf. lavaan R package). Can be one of the
following: “ML” for maximum likelihood, “GLS” for (normal theory)
generalized least squares, “WLS” for weighted least squares (sometimes
called ADF estimation), “ULS” for unweighted least squares, “DWLS” for
diagonally weighted least squares, and “DLS” for
distributionally-weighted least squares. These are the main options that
affect the estimation. For convenience, the “ML” option can be extended
as “MLM”, “MLMV”, “MLMVS”, “MLF”, and “MLR”. The estimation will still
be plain “ML”, but now with robust standard errors and a robust (scaled)
test statistic. For “MLM”, “MLMV”, “MLMVS”, classic robust standard
errors are used (se=“robust.sem”); for “MLF”, standard errors are based
on first-order derivatives (information = “first.order”); for “MLR”,
Huber-White robust standard errors are used
(se=“robust.huber.white”). In addition, “MLM” will compute a
Satorra-Bentler scaled (mean adjusted) test statistic
(test=“satorra.bentler”), “MLMVS” will compute a mean and variance
adjusted test statistic (Satterthwaite style)
(test=“mean.var.adjusted”), “MLMV” will compute a mean and variance
adjusted test statistic (scaled and shifted) (test=“scaled.shifted”),
and “MLR” will compute a test statistic which is asymptotically
equivalent to the Yuan-Bentler T2-star test statistic
(test=“yuan.bentler.mplus”). Analogously, the estimators “WLSM” and
“WLSMV” imply the “DWLS” estimator (not the “WLS” estimator) with robust
standard errors and a mean or mean and variance adjusted test statistic.
Estimators “ULSM” and “ULSMV” imply the “ULS” estimator with robust
standard errors and a mean or mean and variance adjusted test statistic.
|
| ML likelihood approach | Only relevant for ML estimation. If “Wishart”, the Wishart likelihood approach is used. In this approach, the covariance matrix has been divided by N-1, and both standard errors and test statistics are based on N-1. If “Normal” or “Automatic”, the normal likelihood approach is used. Here, the covariance matrix has been divided by N, and both standard errors and test statistics are based on N. |
| Missing values |
Handling of missing values. listwise delete rows with
missing. FIML for ‘full information’ maximum likelihood
estimation. FIML (including fixed x) for ‘full information’
maximum likelihood estimation that includes also the fixed exogenous
variables. Two stages (robust) uses a two stage appraoch
whereas Pairwise deletes case pairwise before computing the
covariances between variables.
|
| Univariate | Score test (or Lagrange Multiplier test) for fixed or constrained parameters in model. |
| Cumulative | Score test for all fixed or constrained parameters in model (cumulative tests). |
Type of models
Users familiar with lavaan or with lavaan documentations
may want to distinguish between different types of models, namely,
cfa (confirmatory factor analysis), sem
(structural equation models) and growth (individual growth
models). These models enjoy the same underlying estimation procedure,
but they are different in the options passed to lavaan. In
SEMLj, one can obtain the same results
as sem and cfa setup using the default options
settings (unless you know you need some change). For growth
model type, one needs to set Intercepts->Observed variables to false
(unselect it), and Intercepts->Latent
variables to true (select it) in the Parameters Options panel, and you’re a happy
camper.
Model parameters
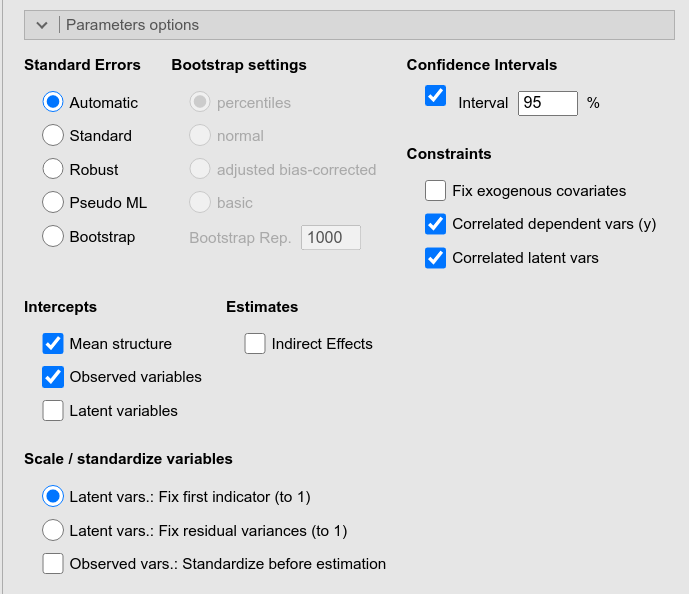
This panel offers many options to be passed to lavaan
for specify parameters charateristics, such as how to compute their
standard errors, their confidence intervals, or how to fix scales of
latent variables.
In addition, it offers the Indirect Effects option which computes and estimate all possible indirect effects (from exogenous to endogenous variables) present in the model.
The options are:
| Standard errors | Standard error method. |
| Bootstrap C.I | Choose the confidence interval type (“perc” - percentiles [default], “bca.simple” - adjusted bias-corrected, “norm” - normal, “basic” - basic). |
| Bootstrap Rep. | The number of bootstrap samples for estimating confidence intervals. |
| Parallel Computation | use parallel computation for bootstrap C.I |
| Estimates |
TRUE or FALSE (default), show confidence
intervals for estimates
|
| Betas |
TRUE or FALSE (default), show confidence
intervals for standardized estimates
|
| Confidence level | A number between 50 and 99.9 (default: 95) specifying the confidence interval width for the parameter estimates. |
| Fix exogenous covariates |
If TRUE, the exogenous covariates are considered fixed
variables and the means, variances and covariances of these variables
are fixed to their sample values. If FALSE, they are
considered random, and the means, variances and covariances are free
parameters. If “default”, the value is set depending on the mimic
option.
|
| Dependent vars (y) |
TRUE (default) or FALSE, TO ADD
|
| Latent vars |
TRUE (default) or FALSE, is the negation of
orthogonal option
|
| Mean structure | If TRUE, the means of the observed variables enter the model. Required for calculating the intercepts of the estimates. |
| Observed variables |
TRUE or FALSE (default), calculate intercepts
for observed variables
|
| Latent variables |
TRUE or FALSE (default), calculate intercepts
for latent variables
|
| Indirect Effects | compute the indirect effect by finding all paths from exogenous to endogenous variables, and multiply the coefficients along the path. |
| Scale / stand. latent vars. |
If Fix first indicator (default), the factor loading of the
first indicator is set to 1.0 for every latent variable. If
Fix residual variances, the metric of each latent variable
is determined by fixing their (residual) variances to 1.0. If there are
multiple groups, Fix residual variances is chosen and
“loadings” is included in the group.label argument, then only the latent
variances i of the first group will be fixed to 1.0, while the latent
variances of other groups are set free.
|
| Observed vars.: Standardize before estimation | If set, all observed variables are standardized before entering the analysis. |
Multi-group Analysis
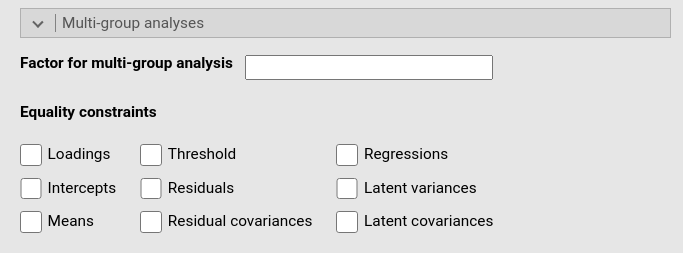
This panel allows for specifying a categorical variable (factor) defining groups of multi-group analysis. One simply writes the name of the factor variable in the Factor for multi-group analysis field.
Once a multi-group analysis is set, one can choose which parameter to fix as equal across groups.
The options are:
| Loadings | constrain the factor loadings to be equal across groups (when conducting multi-group analyses) |
| Intercepts | constrain the intercepts of the observed variables to be equal across groups (when conducting multi-group analyses) |
| Means | constrain the intercepts / means of the latent variables to be equal across groups (when conducting multi-group analyses) |
| Threshold | constrain the sample-based thresholds (means for continuous observed mvariables) to be equal across groups (when conducting multi-group analyses) |
| Residuals | constrain the residual variances of the observed variables to be equal across groups (when conducting multi-group analyses) |
| Residual covariances | constrain the residual covariances of the observed variables to be equal across groups (when conducting multi-group analyses) |
| Regressions | constrain all regression coefficients in the model to be equal across groups (when conducting multi-group analyses) |
| Latent variances | constrain the (residual) variances of the latent variables to be equal across groups (when conducting multi-group analyses) |
| Latent covariances | constrain the (residual) covariances of the latent variables to be equal across groups (when conducting multi-group analyses) |
Multilevel Analysis
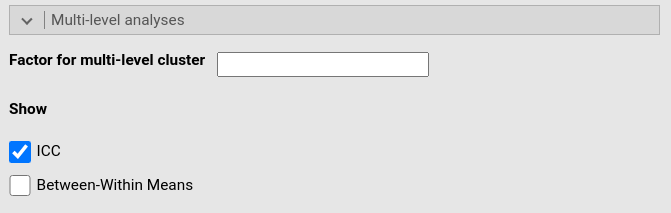
This panel allows specifying a categorical variable (factor) defining cluster variable of multgroup analysis. One simply writes the name of the factor variable in the Factor for multi-group analysis field.
Once a cluster is set, the right syntax for specifying the multilevel model should be set in the syntax panel. See deailts here.
Output Options
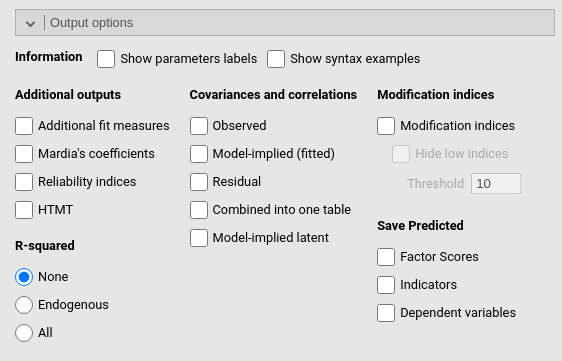
This panel allows requesting all sort of additional information about the model results, such as the \(R^2\)’s, the implied and residual variances and covariances, and the modification indexes.
It is worth mentioning the option to save the predicted and residuals in the dataset. The Predicted option saves, for each participant, the model based predicted values of the observed variables involved in the measurement of purely endogenous dependent variables. The predicted values computation is based on Rooij et al. (2022), and the implementation is adpted from github SEMpredict
The other options are:
| Show parameters labels | show the labels of the parameters in the model |
| Show syntax examples | show examples of the lavaan model syntax |
| Show Lavaan Options | show options of the lavaan model |
| Additional fit measures | show additional fit measures (e.g., CFI, TLI, etc.) |
| Mardia’s coefficients | show Mardia’s coefficients for multivariate skewness and kurtosis |
| Reliability indices | shows additional reliability indices |
| HTMT |
TRUE or FALSE (default), show
Heterotrait-monotrait (HTMT) ratio of correlations as reccomended by
Henseler, Ringle, and Sarstedt (2015)
|
| R-squared |
compute R-squared for all endogenous variables (Endogenous)
or for all variables in the model (All). None
for no R-squared.
|
| Observed | show the covariances and correlations between the observed values of the (manifest) variables |
| Model-implied (fitted) | show the covariances and correlations between the model-implied / fitted values of the (manifest) variables |
| Residual | show the covariances and correlations between the residuals of the (manifest) variables. |
| Combined into one table | combine the (up to) three covariance / correlation tables into one table (i.e., showing observed, model-implied and residual values for each variable combination underneath each other) |
| Model-implied latent | show model-implied latent covariances |
| Modification indices | show modification indices for if certain terms were removed from the model |
| Hide low indices | hide modification indices below a certain threshold. |
| Threshold | Numeric (default: 10), set a threshold the modification indices must exceed in order to be shown. |
| Factor Scores |
this option saves, for each participant, the latent variables predicted
values based on the latent variables estimation. The values are obtained
with lavaan::lavPredict(..,type="lv"). See Rosseel (2012) for
details. The values are basically factor scores.
|
| Indicators |
this option saves, for each participant, the observed variables
predicted values based on the latent variables estimation. The values
are obtained with lavaan::lavPredict(..,type="ov"). See
Rosseel (2012)
for details
|
| Dependent variables | this option saves, for each participant, the model based predicted values of the observed variables involved in the measurement of purely endogenous dependent variables. The predicted values computation is based on Rooij et al. (2022), and the implementation is adpted from github SEMpredict |
The name of the new variables saved in the datasheet are indicated in
a footnote of the Info Table.
Path Diagram
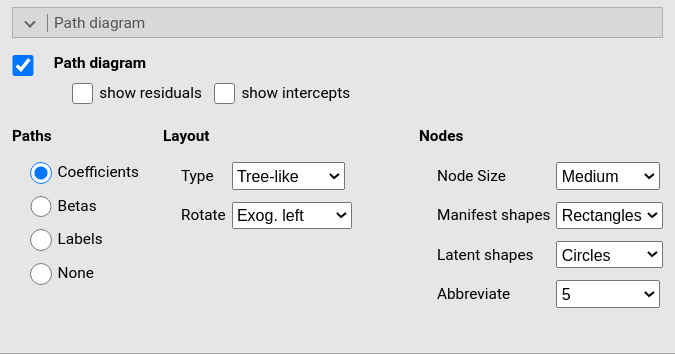
This panel produces the path diagram graph and allow for some change in shapes and orientation of the diagram. The digram is produced using R semPlot module, see also (Epskamp 2015)
The options are:
| Path diagram | produce a path diagram. |
| show residuals | show the residuals (for the observed variables). |
| show intercepts | show the variable intercepts. |
| Paths Labels | Choose the values shown along the paths. |
| Type |
Choose the layout of the path diagram: Tree,
Tree2,Circle,Circle2 ,
Spring).
|
| Rotate | Choose the rotation of the path diagram in terms of placement of the exog. variables. |
| Node Size | Choose the node size for the variables (default: “medium”). |
| Manifest shapes | Choose the shape for the manifest variables (default: “rectangle”). |
| Latent shapes | Choose the shape for the latent variables (default: “circle”). |
| Abbreviate | Choose the length (characters) of the variable name abbreviations (default: 5). |
Examples
Some worked out practical examples can be found here
Comments?
Got comments, issues or spotted a bug? Please open an issue on SEMLj at github or send me an email